Epigenetic Analysis and CRISPR/Cas9 Genome Editing of the Novel Osteoarthritis Susceptibility Locus residing at the PLEC gene
ABSTRACT NUMBER: NESS_13
AUTHORS
Antony K Sorial1, Ines MJ Hofer1, Maria Tselepi1, Eleanor Parker1, Sarah J Rice1, David J Deehan2 and John Loughlin1
AFFILIATIONS
1) Institute of Genetic Medicine, Newcastle University, 2) Freeman Hospital, Newcastle Upon Tyne
CORRESPONDING AUTHOR CONTACT EMAIL
ku.ca.lcn@lairoS.ynoT
MAIN ABSTRACT TEXT
Introduction
Osteoarthritis (OA) is highly polygenic and OA associated single nucleotide polymorphisms (SNPs) are generally non-coding and can function epigenetically, such as DNA methylation changes that alter gene expression. We previously demonstrated that the OA SNP rs11780978 acts as a methylation quantitative trait locus (mQTL) in cartilage, which correlates with reduced expression of PLEC. This gene encodes Plectin, a cytoskeletal protein involved in cellular responses to loading. It has not previously been implicated in OA and here, we set out to expand our molecular characterisation of this OA signal.
Methods
rs11780978 mQTL analyses were undertaken in a range of patient tissues by genotyping and bisulfite-based pyrosequencing. PLEC expression was measured by qPCR. PLEC deletion and knockdown in immortalised adipocyte-MSCs (iAD-MSCs, ATCC-ASC52telo) was performed using CRISPR/Cas9 and lentiviral transduction, with the cells subjected to chondrogenesis.
Results
The mQTL was replicated in cartilage, and was discovered to be active in infrapatellar fat pad, peripheral blood and synovium (Fig. 1A). In synovium, as in cartilage, the risk allele of rs11780978 correlated with reduced PLEC expression. Deletion and knockdown of PLEC was confirmed (Fig. 1B and 1C). Effects on chondrogenesis are currently being determined.
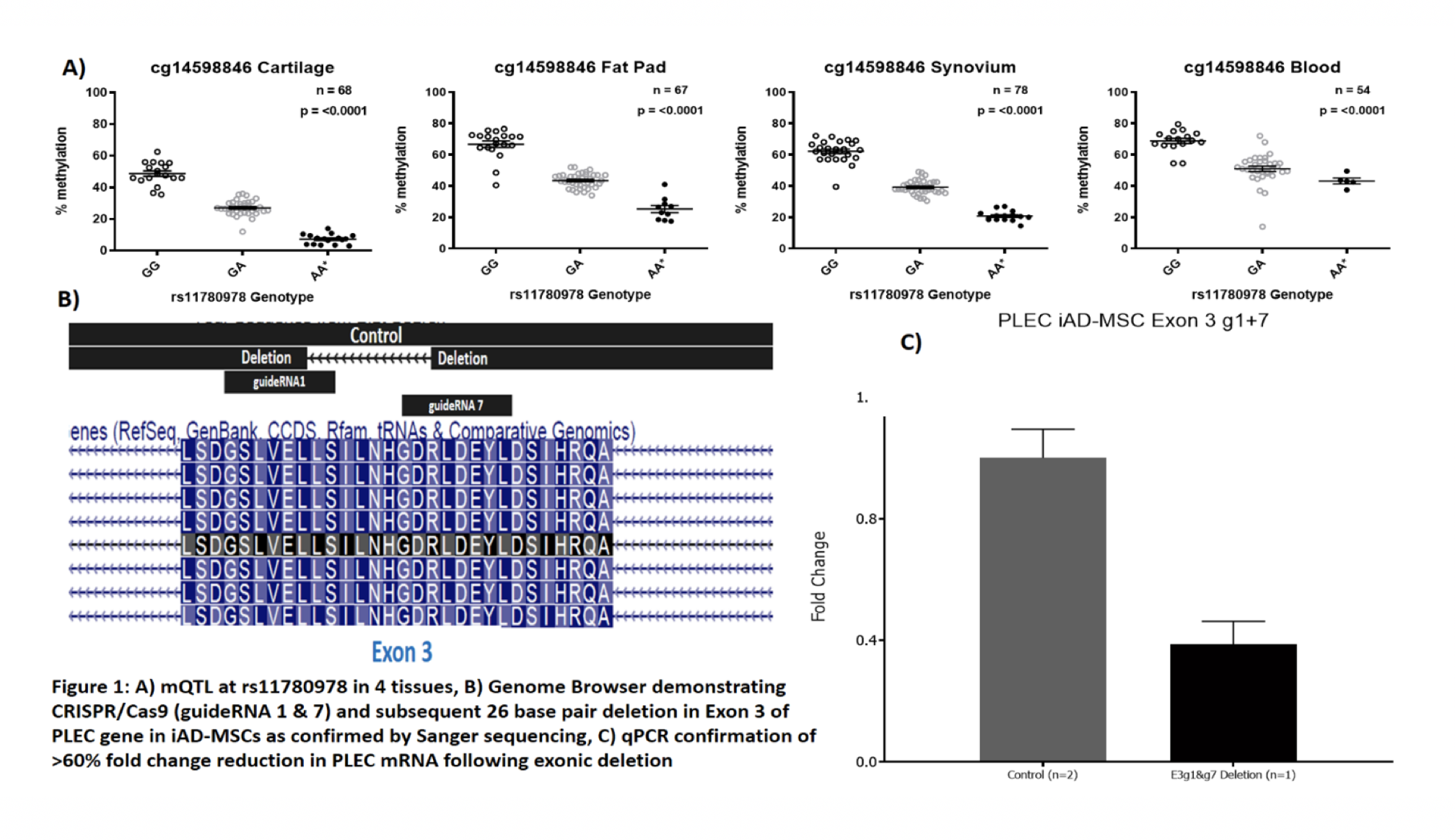 Conclusions
Conclusions
The mQTL has effects in chondrocytes and in non-chondrogenic cells, highlighting the need to investigate multiple cell types when functionally characterising OA genetic risk. PLEC knockdown, which models the genetic deficit has been undertaken and effects on phenotype will shortly be known. We have developed tools to investigate OA risk and can apply these for future OA molecular studies.
